The Maranas group is working on the development of algorithmic and, in particular, optimization techniques to support the analysis and redesign of biological systems at different scales.
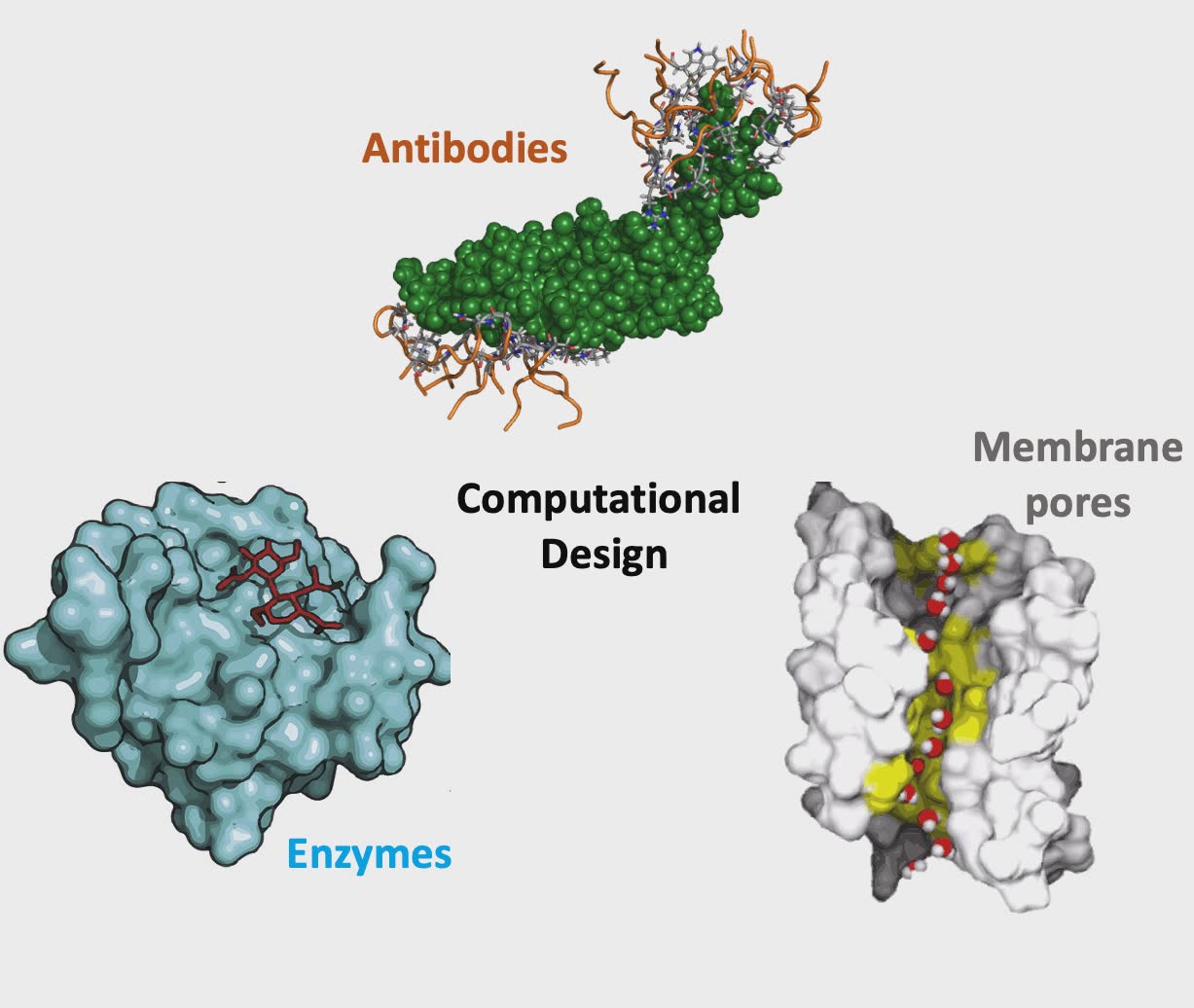
At the protein level, we are interested in computationally inferring what amino acid compositions are likely to yield i) proteins or antibodies with targeted binding affinities, ii) enzymes with improved stability, specificity and activity for specific biotransformations, and iii) protein pores that allow for tailored separations. To this end, we rely on biophysics inspired force fields such as CHARMM and Rosetta to quantify molecular interactions.
At the metabolic network level, we are pursuing methods for automating the generation, curation, and correction of genome-scale models of metabolism. We are also interested in generating isotope mapping models to support metabolic flux elucidation using MFA. In addition, we are working towards developing computational tools to help decide how to engineer (i.e., through gene knock-in/out/up/down(s)) biological production systems. A unifying feature of these seemingly disjoint research targets is the need to systematically search through many network configurations, amino acid compositions, protein structures, etc. and identify the "best" one. To this end, the development of efficient theoretical, algorithmic, and computational techniques for arriving at relevant as well as theoretically sound results while maximizing computational efficiency is pursued.
Professor Maranas is also a member of Faculty of the Operations Research Program. Operations Research (OR) is the use of scientific methodology in the formulation, analysis, and solution of problems in decision making. It draws on techniques from many fields, including economics, mathematics, and engineering. Students with a strong interest in operations research techniques can apply for an Operations Research Dual-title degree, such as a Dual Master degree in OR, Dual Ph.D. degree in OR, and Minor Ph.D. degree in OR. Interested students must submit an application for admission to the Chair of the OR Program and meet the necessary course requirements. Relevant information can be found at: https://www.or.psu.edu/.